Assembling Raspberry on Raspberry Pi
A group
of scientists has managed to assemble the hefty 400 million base pairs
of the raspberry genome
on . . . the eponymous Raspberry
Pi, a mini-computer with only 512MB of RAM. The prowess was meant to
demonstrate the dramatically low memory footprint of a new generation
of software designed by GenScale, a bioinformatics project team at
Inria research center, in Rennes, Brittany, France. This technology
will be made available to biologists through GATB, a
soon-to-be-released toolbox dedicated to genome assembly.
|
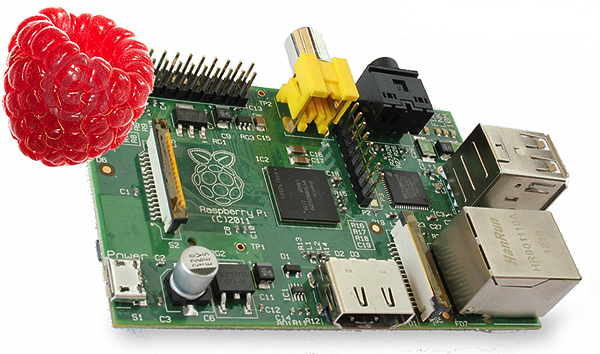 Featuring a 800 MHz CPU and 512 MB RAM, the Rasperry Pi is less powerfull than a smartphone.
Featuring a 800 MHz CPU and 512 MB RAM, the Rasperry Pi is less powerfull than a smartphone.
It weighs merely 45 g., operates under Linux and costs only $35. |
Sequencing a genome has become a rather routine operation in recent
years due to new generation sequencers (NGS). These relatively
inexpensive machines generate heretofore unseen volumes of data: DNA
molecule is recorded as millions of strings in the ‘A C G
T’ alphabet. Yet, a problem remains: the outpout is not readily
exploitable. It is like a puzzle made of short fragments
—genomic reads— that need to be compared and
assembled in order to reconstruct the sequence. Such a task is the
raison d'être of specific applications called assemblers. Alas these
tools come with a drawback: one needs a cluster to run them. In a
context of data deluge, assembly is thus emerging as the bottleneck in
biologists' workflow.
“Memory footprint is really the crux of the matter
here,” sums up Dominique
Lavenier, head
of GenScale (1),
a research team that has taken upon itself to redesign the
underpinning algorithms of such tools. As a result of a lengthy effort
of code optimization, scientists are coming up with a suite of
applications that run with little RAM
(2). “Roughly 20 to 50 times less
memory compared to previous tools in the field.” Such
quantum leap could prove a game-changer for biologists.
“They will be able to assemble on desktops instead of
clusters.”
|
In order to demonstrate the RAM frugality of their novel algorithms,
the researchers went as far as trying to run one application on
the Raspberry Pi, a famous
credit-card-sized rudimentary computer sporting a paltry 512MB of
memory. The first major experiment was conducted on C.
Elegans, a microscopic roundworm whose 100 million
base pairs genome was assembled in about 19 hours. “It
didn't work the first time, which prompted us to undertake further
code optimization. But in the end of the day, we
succeeded.” Credit is due to
postdoc Guillaume Collet who
managed to implement the software on so little a device Shortly
therafter the idea popped up of trying to assemble the Raspberry
genome and its daunting 400 million base pairs on the eponymous
mini-computer. A professor at Brigham Young University in
Utah, Joshua
Udall graciously supplied the sequence dataset. The assembly then
took a solid week, but it worked, opening the door to more convenient
assembly on run-of-the mill desktop computers.
Instrumental to this effort were two PhD students.
“ Rayan
Chikhiand Guillaume Rizk
were the ones who brought up the seminal
ideas(3). Three computer scientists at
Paris-Est
Marne-la-Vallée University also
joined in, adding a very valuable contributio(4) to
our code optimization. After a while, it became apparent that their
research had laid the foundation for a real software that could be of
service to the scientific community.” At this juncture, the
French research
agency ANR
stepped up to the plate with a €180,000 funding meant to help
morphing what was still a scientific prototype into a fully
operational software. The hope is also to foster a technology transfer
to the industry. Interestingly, the funding by ANR enabled Genscale to
recruit an engineer who completely refashioned the software
structure. “Erwan Drezen put a strong emphasis on building
libraries as bases for all our applications. In hindsight, this proved
very relevant as creating specific tools from these libraries now only
requires a few days of work.”
|
Many Tools in the Box
Releasded under AGPL license, the GATB
suite comprises a variety of applications
that address the distinct phases of genome assembly. Namely: k-mer
counting, contig construction and scaffolding. These successive steps
could be described as: assembling small parts, bigger chunks and the
final puzzle.
For k-mer counting, biologists will use DSK and then switch
to BLOOCOO,
which is a read error corrector. Errors being contingent on the
different sequencers on the market, several NGS-specific correctors
will actually be added to the toolbox. Next in line stands Minia,
an application designed for contig construction, which incidentaly
happens to be the most costly phase. As a stand-alone tool, Minia has
already been downloaded 10,000 times. “At the moment, we are working on
the scaffolding step as well as on filling the gaps between the
scaffolds. Other applications will follow, including MapSembler which
is
an assembler targeted on given regions of interest.”
|
Full toolbox profile at http://gatb.inria.fr
|
The libraries will be part of the toolbox, thus enabling
computer programmers to swiftly compose their own high-throughput
low-memory applications. “And
we very much encourage them to do so,
Dominique Lavenier concludes. Indeed,
assembling the genome of a bacterium or a
mammal calls for different approaches and different tools. Therefore,
our vision is that genomic assembly should rely on a multitude of
custom-made assemblers targeted for specific species.”
Publication: E. Drezen, G. Rizk, R. Chikhi, C. Deltel, C. Lemaitre, P. Peterlongo, D. Lavenier, GATB: Genome Assembly & Analysis Tool Box, Bioinformatics, 2014
(link)
- - - -
Notes:
(1)
Scalable Optimized and Parallel Algorithms for Genomics: GenScale is
a
joint project team of Inria/ENS Cachan/Rennes 1 University, common to
Irisa (UMR CNRS 6074). Guillaume Collet is a member of Dyliss, a joint
project team of Inria/CNRS/Rennes 1 University, common to Irisa.
(2) Random Access Memory. RAM is used as temporary storage for the
applications.
(3)
Read: R. Chikhi, G. Rizk. Space-efficient
and exact de Bruijn graph
representation based on a Bloom filter, Algorithms for
Molecular
Biology 2013, 8:22.
(4) Read: K.l Salikhov, G. Sacomoto,
G. Kucherov,
Using
Cascading Bloom Filters to Improve the Memory Usage
for de Brujin Graphs, Algorithms in Bioinformatics,
Lecture Notes in
Computer Science, Volume 8126, 2013, pp 364-376
|
|
|